

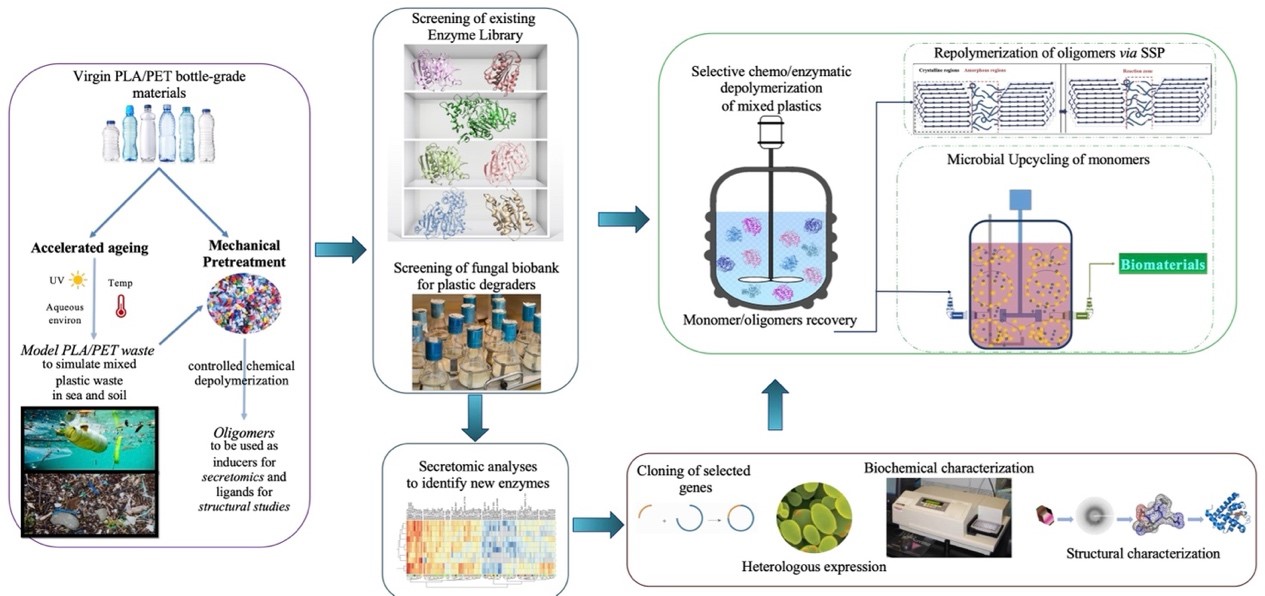
The overall aim of EnZyReMix is the development of novel bioprocesses capable of selectively breaking down and thus separating synthetic polymers, such as PLA and PET, for the chemoenzymatic recycling of complex packaging waste streams, such as mixed plastics.
Schematic of the EnZyReMix Project
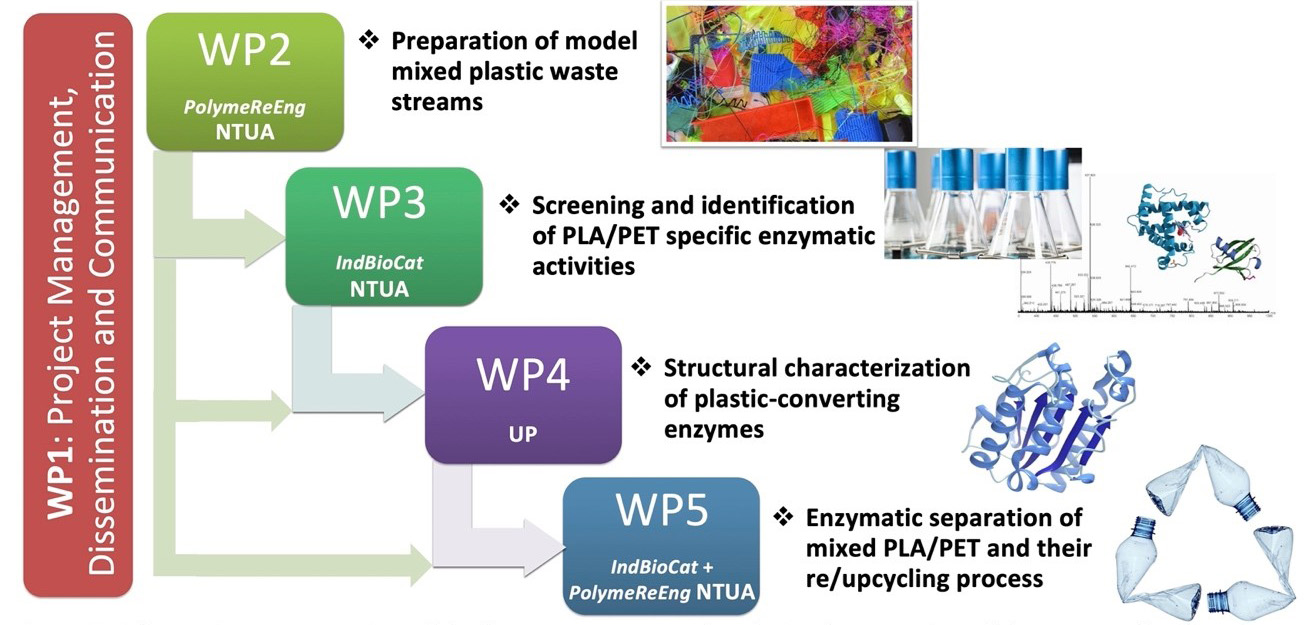
The work plan of EnZyReMix consists of 5 closely interconnected work packages (WPs), implementing an integrated holistic approach for the discovery of novel plastic-modifying enzymes for the separation/selective depolymerization of mixed polyesters waste, their functional and structural characterization and finally, a proof-of-concept re- and up-cycling of the waste plastic materials.
Schematic representation of the five WPS necessary for the implementation of the Project
WP1 includes the coordination and management of the proposed project and all the involved actions, along with dissemination and communication activities. As a first R&D step, different PLA/PET mixtures will be treated under conditions that simulate soil and sea environments producing model mixed plastic wastes (WP2), while controlled chemical depolymerization will be carried out to produce corresponding oligomers for structure-function relationship studies for WP4, as well as inducers for Secretomics studies for WP3. In WP3, an in-house library of enzymes, as well as commercially available enzyme preparations will be screened for their ability to modify virgin or weathered PET and PLA polymers derived from WP2. Bioinformatic mining of secretomic data generated from the most competent strains isolated from areas polluted with plastics and petrochemicals provided by UB, will be used for the discovery of serine hydrolases among other enzymatic activities. WP4 is dedicated to the structure elucidation of targeted biocatalysts, which together with the biochemical data from WP3, will unveil the unique catalytic properties of plastic-modifying enzymes. The acquired information from these WPs will be applied in the enzymatic separation of PLA/PET mixtures and the repolymerization of the resulting monomers/oligomers, in addition to their upcycling through microbial fermentation towards new biomaterials as proof-of-concept (WP5).
Plastic pretreatment
Solvolysis of PLA and PET bottle-grade virgin polymers aiming to create oligomers to be used as induction media for the secretion of PLA and PET specific enzymes from target fungal strains through proteomics, as well as substrates for co-crystallization studies.
Artificial lab weathering of PLA and PET materials that can be considered of similar degradation extent as the ones improperly disposed in soil and sea. Aged materials at different timeframes will be thoroughly characterized to construct the most critical structure/properties relationships for the subsequent enzymatic degradation.

Q-Lab QUV Weathering Chamber for artificial weathering of PLA and PET
Screening of characterized and discovery of novel enzymes for PET and PLA
 Screening of in-house library for polyesterases: IndBiocat enzyme library consists of over 15 polymer-active enzymes and will be screned with virgin PET and PLA materials classifying each enzyme as a PETase or PLAase.
Screening of in-house library for polyesterases: IndBiocat enzyme library consists of over 15 polymer-active enzymes and will be screned with virgin PET and PLA materials classifying each enzyme as a PETase or PLAase.
 Discovering novel fungal enzymes through proteomics: Oligomers deriving from plastic pretreatment will be utilized as enzyme inducers in fungal cultures and secreted proteins will be identified by nanoLC-MSMS to discover enzymes with activity on PET or PLA.
Discovering novel fungal enzymes through proteomics: Oligomers deriving from plastic pretreatment will be utilized as enzyme inducers in fungal cultures and secreted proteins will be identified by nanoLC-MSMS to discover enzymes with activity on PET or PLA.
Structural characterization of selected enzymes (WP4)
Novel enzymes that exhibit interesting properties but are also stable and can be adequately purified, will be subjected to structural studies by X-ray crystallography.

X-ray crystallography equipment
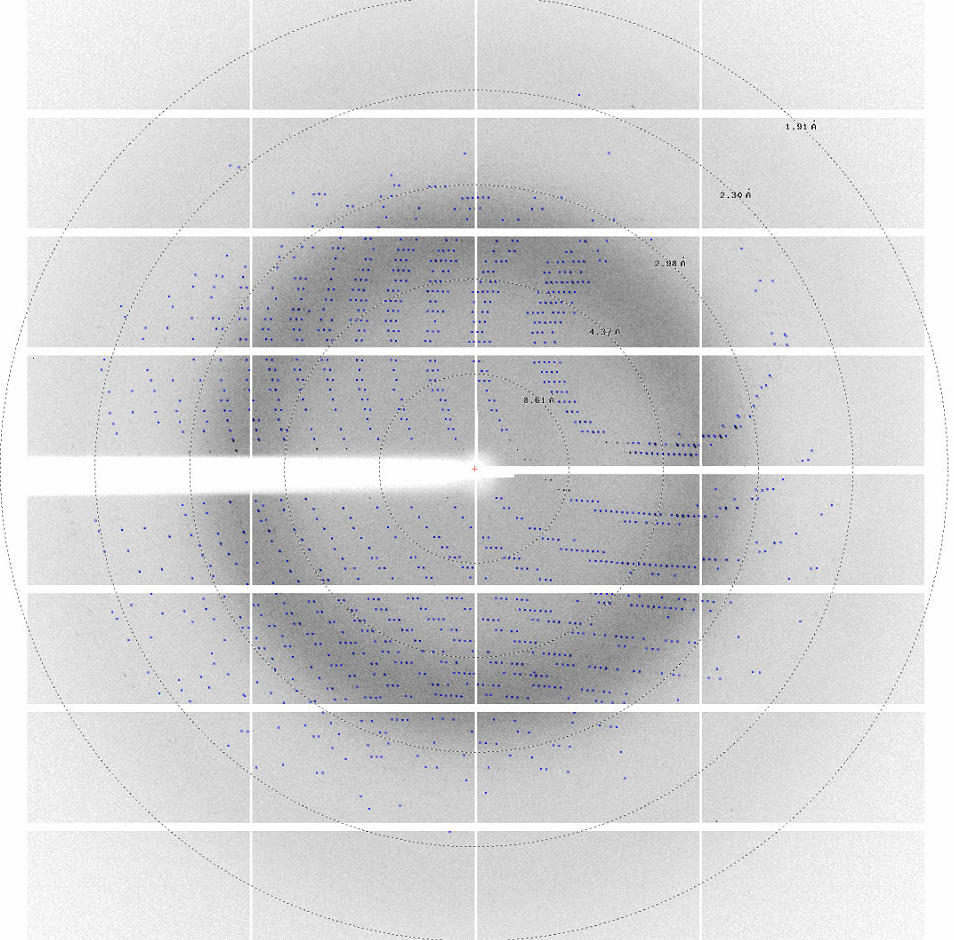
 To get deeper insight into enzyme mechanism, the structures of interesting enzymes in complex with substrate analogues or inhibitors will be targeted. Model PET compounds such as terephthalic acid (TPA), monohydroxyethyl terephthalate (MHET) and other PET oligomers are available at the NTUA group, while the chemical decomposition of PLA by PolymeReEng group will allow the use of mixtures of PLA oligomers for co-crystallization studies. These structures will be used to elucidate the amino acids implicated in substrate binding and catalysis.
To get deeper insight into enzyme mechanism, the structures of interesting enzymes in complex with substrate analogues or inhibitors will be targeted. Model PET compounds such as terephthalic acid (TPA), monohydroxyethyl terephthalate (MHET) and other PET oligomers are available at the NTUA group, while the chemical decomposition of PLA by PolymeReEng group will allow the use of mixtures of PLA oligomers for co-crystallization studies. These structures will be used to elucidate the amino acids implicated in substrate binding and catalysis.
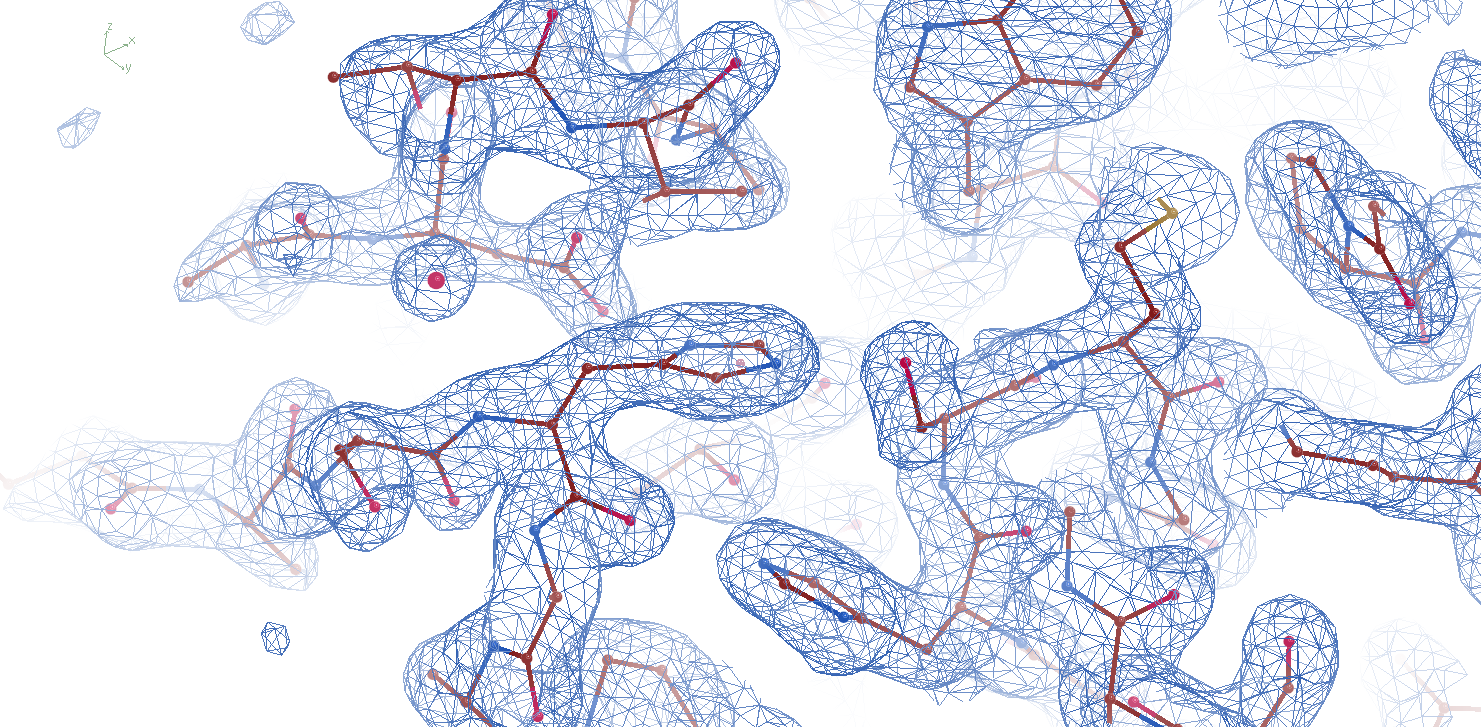
Electron density at the active site of an engineered LCC variant
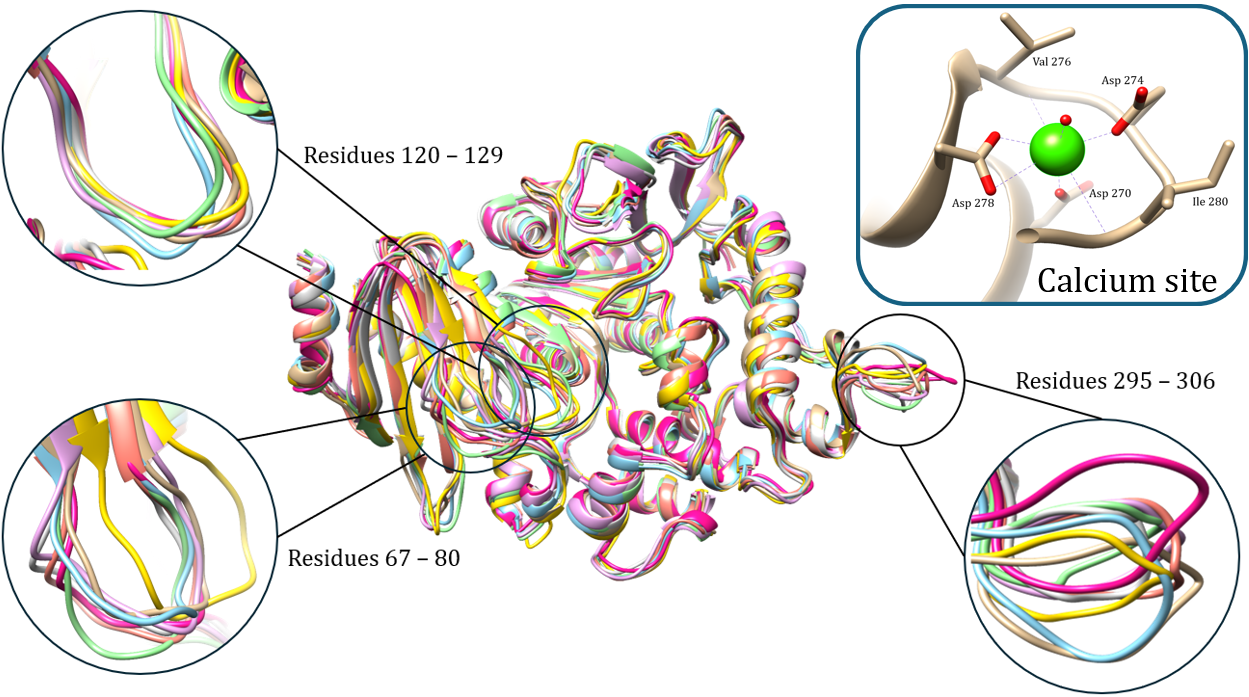
Molecular dynamics simulations to analyze the effect of point mutations on MHET active enzyme
Selective enzymatic depolymerization of PLA/PET mixtures and recycling or upcycling of monomers (WP5)
Exploiting the strong specificity of targeted biocatalysts that selectively depolymerize one of the two polyesters, while maintaining the second one without any severe degradation. A mild hydrolysis process to separate PLA/PET mixtures will be employed aiming in the sustainable separation and subsequent oligomerization or complete depolymerization of both PLA and PET streams. Depending on the re- or upcycling strategy followed, the combination with chemical hydrolysis for the degradation of remaining polymer will be aimed to recover the monomers/oligomers.
Recycling of separated polymer streams will be performed by repolymerization of the resulting oligomers through melt and/or solid state polymerization (SSP).
Upcycling of monomers will be done using P. putida and K. medellinensis strains provided by IMGGE-UB towards the production of biomaterials, such as mcl-PHA and bacterial nanocellulose.


Overall, the project generated 20 dissemination items: 3 peer-reviewed open-access publications, 15 conference participations and 2 PDB enzyme structure depositions. The crowds reached were scientists, engineers and industry stakeholders related to Polymer Science and Material Engineering, Chemical Engineering, Enzymology, Biocatalysis and Biotransformation. Through these activities we managed to reach over 31000 academic and industrial stakeholders.
Papers
Polyester-derived monomers as microbial feedstocks: Navigating the landscape of polyester upcycling.
- Biotechnology Advances, 82 (2025), 108589
View paper »
Protein Engineering for Industrial Biocatalysis: Principles, Approaches, and Lessons from Engineered PETases.
- Catalysts 2025, 15(2), 147
View paper »
Structural investigation of an engineered feruloyl esterase with improved MHET degrading properties.
- BioRxiv
View paper »
Conference Presentations
Harnessing the catalytic potential of a ferulic acid esterase for MHET hydrolysis.
- LignoBiotech 2024, Toulouse, France
View abstract » View poster »
Mimicking the enzymatic plant cell wall hydrolysis machinery for the degradation of polyethylene terephthalate.
- LignoBiotech 2024, Toulouse, France
View abstract » View poster »
Reviving mixed plastic waste through chemoenzymatic recycling.
- Polymers 2024: Polymers for a Safe and Sustainable Future, Athens, Greece
View abstract » View poster »
Valorization of Monomeric Compounds Originating from Synthetic Polymers by the Heterotrophic Microalgae Crypthecodinium Cohnii.
- Panhellenic Conference of Chemical Engineering 2024, Thessaloniki, Greece
View abstract » View poster »
Investigation of Structural Determinants of Plastics Degrading Enzymes via X-ray Crystallography and Molecular Docking.
- Panhellenic Conference of Chemical Engineering 2024, Thessaloniki, Greece
View poster »
Screening Biocatalysts for Targeted Polyester Degradation.
- Microbetech 2025, Ljubljana, Slovenia
View poster »
Structural basis for enhanced MHET degrading activity of engineered feruloyl esterase.
- EMBO Workshop: Computational Structural Biology, Heidelberg, Germany
View abstract » View poster »
Engineering Polymers for a Beyond-the-bin Strategy to Recycle Mixed Plastic Waste.
- EPF European Polymer Congress 2025, Groningen, The Netherlands
View abstract » View poster »
Biotechnological valorization of PLA hydrolysates via microbial PHA synthesis by Pseudomonas spp.
- Joint International Conference of Mikrobiokosmos & CEESME, Thessaloniki, Greece
View abstract » View poster »
Structural insights into the catalytic mechanism of plastic-degrading enzymes.
- Joint International Conference of Mikrobiokosmos & CEESME, Thessaloniki, Greece
View abstract » View poster »
Comparing the secretome response of Aspergillus and Fusarium species on chemically treated plastics.
- ECCS2025-The 3rd International Electronic Conference on Catalysis Sciences, 23–25 April 2025, online
View abstract » View poster »
Screening biocatalysts for the selective enzymatic separation of polyester blends.
- ECCS2025-The 3rd International Electronic Conference on Catalysis Sciences, 23–25 April 2025, online
View abstract » View poster »
Structural study of an engineered leaf branch compost cutinase (LCC) with PET oligomers.
- COST Action CA21162 Working Group meeting, 24-25 April 2025, Athens, Greece
View abstract » View poster »
Structural studies and molecular dynamics simulations of a ferulic acid esterase variant active on PET oligomers.
- COST Action CA21162 Working Group meeting, 24-25 April 2025, Athens, Greece
View abstract » View poster »
Engineering the activity of a thermophilic esterase from Zhizhongheella caldifontis for MHET degradation
- Joint International Conference of Mikrobiokosmos & CEESME, Thessaloniki, Greece
View abstract » View poster »